
Triplex
Intermolecular triplex molecules generated by 3D-NuS are categorized into 2 classes and 8 sub-classes. The two classes are defined according to the orientation of 3rd strand with respect to the purine rich strand and they are either parallel or anti-parallel. Possible nucleotide for these two classes are described in the table below. The 8 sub-classes are defined with respect to the molecular nature of individual strands of triplexes (either DNA or RNA) and are described in the under given table.
Input sequences for strand one and two must contain only pyrimidines and purines respectively. Refer under given figure and documentation for different acceptable combinations of hydrogen bonding patterns, base triplets and class for triplexes. If the categories of the molecule are not defined by user, then default values in respective categories will be considered for molecule generation. Click 'Submit' to generate the PDB file of the triplex molecule. User can view the generated 3D structure in the given molecule viewer or can 'Download' and visualize using other molecule visualizers.
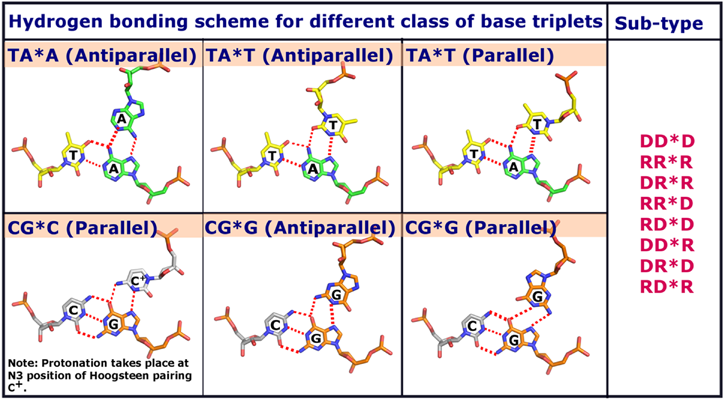
Figure 2. Base triplet hydrogen bonding schemes considered for triplex modeling in 3D-NuS.
Here, D&R represent DNA and RNA respectively.
Let's build the molecule of your choice...
Select appropriate type of triplex from undergiven options.